Long-Non Coding RNAs – Hidden in Plain Sight
In the last 60 years, molecular biologists have focused on studying the 2% of the human genome that contains coding genes. However, the remaining 98% of the genome may hold secrets that have a significant bearing on many critical biological questions and life-threatening genetic disorders. In recent years, thousands of new genes were discovered, hidden in plain sight within the understudied 98% of the genome scientists used to refer to as “junk DNA”. Therefore, the attitude towards “junk DNA” is changing and it is now widely referred to as “non-protein coding genes’ or “non-coding RNA”.
In our lab, we are especially interested in a new group of non-protein coding genes, called long non-coding RNAs (lncRNAs), which share many features with protein-coding genes. The mammalian genome encodes for thousands of lncRNAs, but the reason for their existence is still a big mystery. Why do we have these genes? Do these transcripts play an essential role in cell function? What is the role of lncRNAs in disease, and how can we exploit it to benefit of human health?
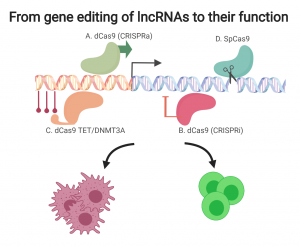
CRISPR-dCas9 opens unprecedented opportunities to study the non-coding genome.
The recent development of CRISPR-Cas9 (clustered, regularly interspaced, short palindromic repeats (CRISPR) and the CRISPR-associated protein 9 (Cas9)) technology has opened new and exciting possibilities for genetic studies. Using CRISPR technology, the DNA sequence can be easily manipulated, for both in vitro and in vivo studies. Endonuclease-deficient Cas9 (dCas9), which has been recently developed, preserves the targeting function of CRISPR without DNA sequence modulation. A particularly useful application for this technology is to add a transcription repression domain or transcriptional activators to dCas9, thus enabling the regulation of “the volume” of gene expression from their endogenous site without interfering with the DNA sequence. This manipulation is termed CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa) and it allows us, for the first time, to perform intensive functional studies of non-coding elements such as lncRNAs, enhancers and the epigenome, in their native genomic contexts.
In our lab, we use high throughput gene-editing tools in an effort to uncover the functional role of non-coding genes during cell differentiation in health and disease.